Hi. I'm following this tutorial (https://developers.refinitiv.com/en/article-catalog/article/using-tick-history-in-r-language-part-3) to use Refinitiv REST API in R. But I think the question should be general and apply to other languages (Python/C etc).
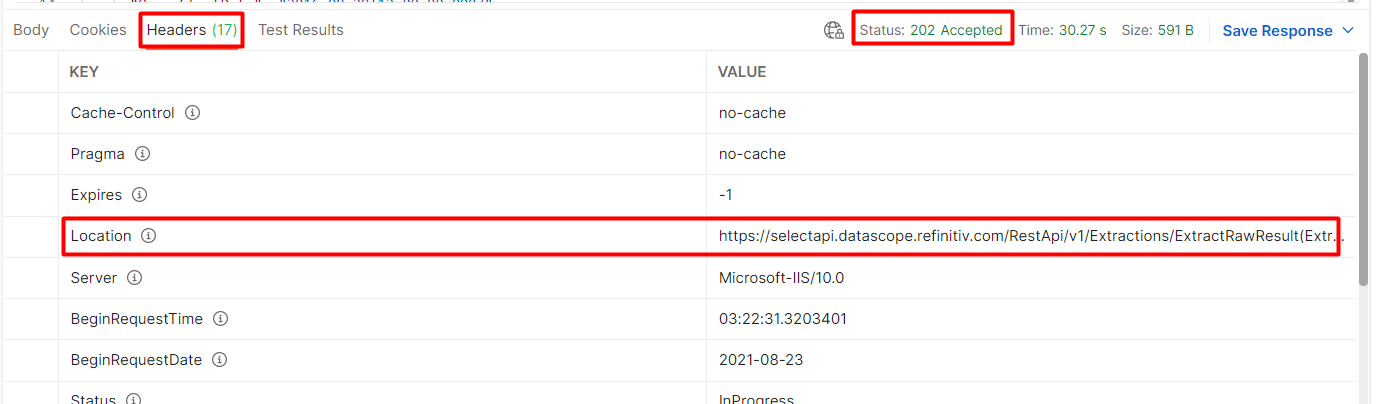
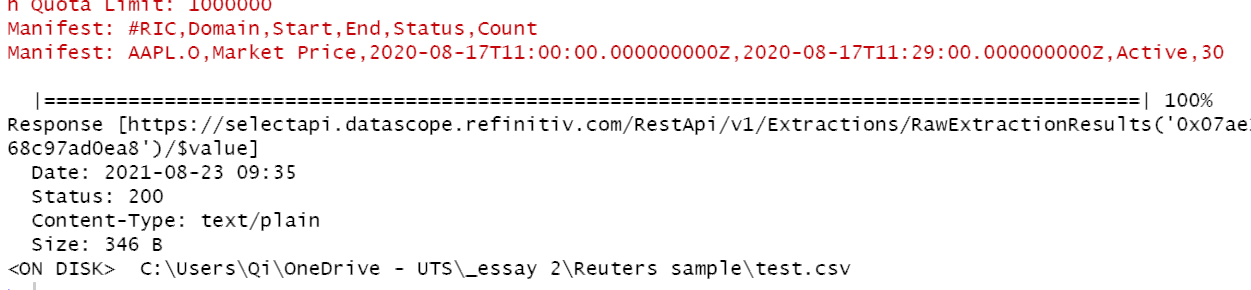
So I'm not able to get results within 30s time-out period so I'm polling the 'get' method. When running:
url <- "https://selectapi.datascope.refinitiv.com/RestApi/v1/Extractions/ExtractRaw"
r <- httr::GET(location,add_headers(prefer = "respond-async",Authorization = token))
I get the error:
Error in handle_url(handle, url, ...) : ccan't find object 'location'
But nowhere in the tutorial mentions how to define 'location'. So can I get some help on this issue? Thank you.